Untangling antimicrobial resistance
Treating bacterial diseases is becoming increasingly difficult, but the Veterinary Diagnostic Laboratory is facilitating more targeted treatments

Treating bacterial diseases is becoming increasingly difficult, but the Veterinary Diagnostic Laboratory is facilitating more targeted treatments
Bacteria, fungi, parasites, and viruses aren’t as simple as they may appear. The microscopic organisms have a talent for evading treatments, which means the defenses we have today are losing power to antimicrobial resistance (AMR). The good news is, by better understanding how certain pathogens tick, and which treatments are most likely to work, scientists can prevent the kind of treatment guesswork that fuels resistance.
The University of Minnesota (UMN) Veterinary Diagnostic Laboratory (VDL) is a hub for such antimicrobial stewardship, especially monitoring livestock diseases. Jerry Torrison, DVM, PhD, DACVPM, director of the VDL, says it’s a combination of top-tier experts, close relationships with real-world stakeholders, and the sheer volume of samples the VDL processes that have made the lab a leader in education, research, surveillance, and management of AMR in the field.
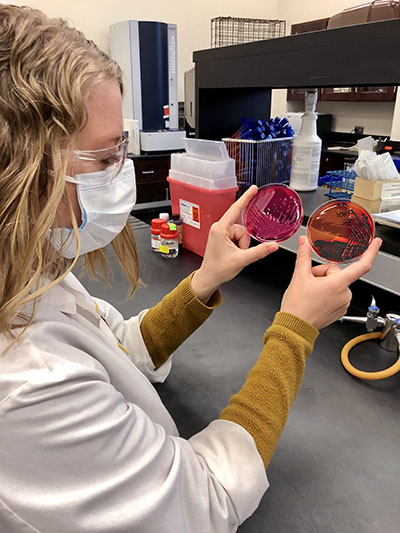
The VDL’s Bacteriology Section performs almost 30,000 bacterial cultures annually, at least 3,000 of which are screened for potential antibiotic resistance. By identifying which antibiotics will likely be able to treat certain strains of a bacterium, and which it’s unlikely to, the VDL helps farmers and veterinarians eliminate unnecessary antibiotic use that contributes to resistance.

The work also fine-tunes vaccines that can specifically target a bacterium variant. Currently, vaccines based on certain isolates are being used to treat and prevent outbreaks of swine disease on individual farms and to inform nationwide disease control efforts.
But the value doesn’t end there.
Most of these samples also are archived for future use, meaning the information locked in these isolates can be used to solve problems that haven’t surfaced yet.
“We have been doing bacterial cultures for over 100 years, so we have a lot of experience,” Torrison says, adding that the lab not only helps practitioners with individual cases but also identifies trends over time. “These trends are only visible because the VDL has built such a deep archive of samples throughout time.”
According to Albert Rovira, DVM, PhD, assistant director of the VDL, cutting-edge technology is allowing the team to pull even more information from both archived and current samples.
“The traditional method of AMR testing allows veterinarians and producers to know right away what antibiotics they can and cannot use. This is routine,” Rovira says. “But when we go more in-depth and investigate the whole genome, we can understand why cases are resistant. We can understand how this resistance travels among bacteria, which scientists need to understand when developing tests and treatments for AMR bacteria.”
This research is solving some of the most pressing problems facing human, animal, and environmental health both today and in the future.
The U.S. Department of Agriculture (USDA) Animal and Plant Health Inspection Service National Animal Health Laboratory Network project is monitoring AMR profiles in animal pathogens, including Salmonella species, E. coli, and Staphylococcus species. Keeping close tabs on each emerging strain allows scientists, regulators, and policymakers to pinpoint which antibiotics are becoming less effective against certain diseases.
The agency is able to do this by aggregating samples from 21 labs throughout the U.S. and Canada, including the VDL, and distilling the information in one centralized location. Every year, they share information on emerging AMR trends with veterinarians, food producers, and other stakeholders who can apply it in the field.
Connie Gebhart, PhD, faculty advisor for bacteriology for the VDL, and her team used advanced genome sequencing to improve control of Streptococcus suis, a potentially zoonotic bacteria that can be transferred from pigs to humans in rare circumstances. To do this, her team collaborated with the Kansas State Veterinary Diagnostic Lab to characterize 208 samples of S. suis. Their goal was to determine which individual genes made a strain more likely to be resistant to certain types of antibiotics. They were able to identify 17 genes that were associated with resistance to at least one of eight commonly used antibiotics.
“AMR is one of the big issues coming forward for humans, animal pathogens, and environmental health and it’s incredibly important for industry, government, and academia to cooperate in their efforts to investigate AMR and how we can control it,” Gebhart says.
Like the USDA, the Food and Drug Administration (FDA) is closely monitoring strains of pathogens that are circulating in the U.S., and how susceptible different strains are to available treatments.
The FDA Center for Veterinary Medicine, Veterinary-Laboratory Investigation and Response Network project has worked to standardize sequencing capabilities of several key labs in the U.S. that are closely involved with the National Antimicrobial Resistance Monitoring System. To do so, the FDA outfitted the VDL with advanced genome sequencing equipment that will make continued collaboration possible.
“Instead of sending samples to the FDA, we will be able to routinely do genome sequencing in-house,” Gebhart says. “It’s also getting us prepared for any outbreaks. If we all use the same equipment, there can be a standardized, all-hands-on-deck approach, and the government can rely on our labs to track and understand future outbreaks.